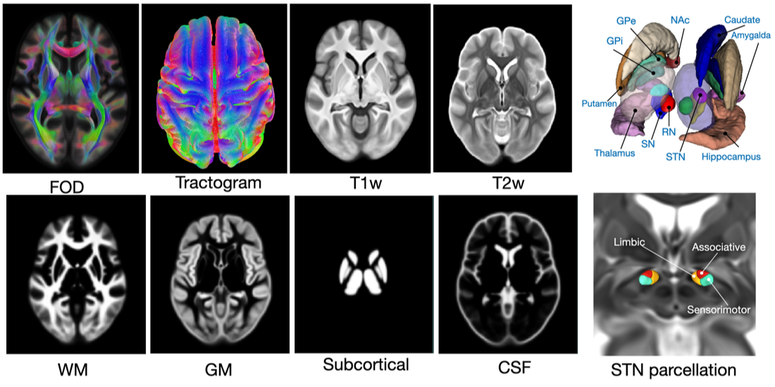
HCP-A422 structural connectomic atlas
This data repository provides a set of multi-contrast structural connectomic brain atlas, constructed using multi-contrast group-wise image registration of 422 Human Connectome Project - Aging group subjects. Besides population-averaged structural MRI template, FOD map, and tractograms, tissue segmentation and functional parcellation of the subthalamic nucleus are also provided.
For further details regarding this dataset, please refer to the following article:
For further details regarding this dataset, please refer to the following article:
- Y. Xiao, J.C. Lau, T.M. Peters, A.R. Khan, "A population-averaged structural connectomic brain atlas of 422 HCP-Aging subjects," Data in Brief, 109513, 2023.
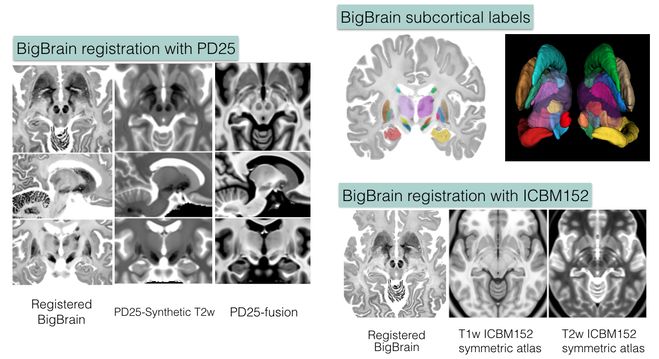
BigBrain co-registration with the MNI ICBM152 and PD25 atlases
The repository contains validated co-registration between the BigBrain dataset (bigbrain.loris.ca) with ultra-high resolution histological brain data and the MNI ICBM152 and PD25 brain MRI atlases. In addition, manual segmentation of 11 pairs of subcortical structures were also provided for all datasets involved in high resolution. The repository provides a bridge between the insights at microscopic and macroscopic scales for the study of the human brain, and is highly valuable for both the research and clinical community.
For further details, please refer to the following article:
For further details, please refer to the following article:
- Y. Xiao, J.C. Lau, T. Anderson,J. DeKraker, D. Louis Collins, T.M. Peters, and A.R. Khan, “An accurate registration of the BigBrain dataset with the MNI PD25 and ICBM152 atlases,” Scientific Data, accepted, 2019.
- 1. Amunts, K. et al.: “BigBrain: An Ultrahigh-Resolution 3D Human Brain Model”, Science (2013) 340 no. 6139 1472-1475, June 2013
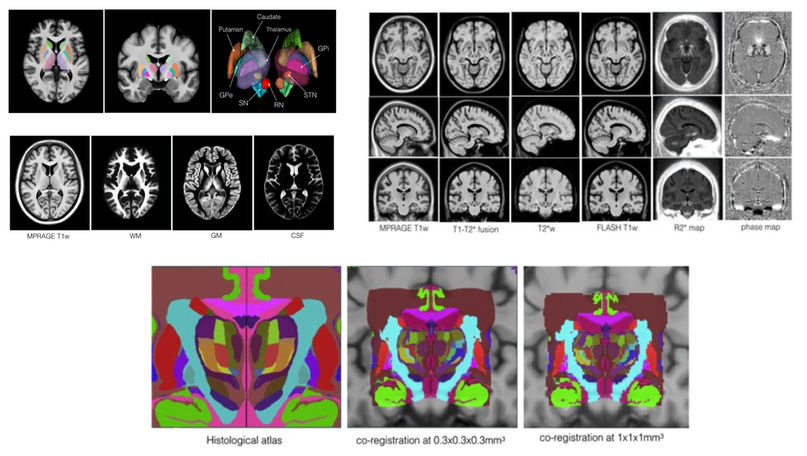
Multi-contrast Parkinson's disease brain atlases (MNI PD25)
The repository presents a collection of multi-contrast MRI templates constructed from 25 Parkinson’s disease patients, along with manually segmented subcortical structures, a co-registered histology-based atlas of 123 structures, and co-registered big brain atlas (https://bigbrain.loris.ca). The repository provides tools for the surgical planning and image-based study of movement disorders. The data repository is shared with CC BY-NC-SA 3.0 license.
For the methods used, and to use the atlas for research purposes, please cite the following articles:
For the methods used, and to use the atlas for research purposes, please cite the following articles:
- Y. Xiao, V. Fonov, S. Beriault, F.A. Subaie, M.M. Chakravarty, A.F. Sadikot, G. Bruce Pike, and D. Louis Collins, “A dataset of multi-contrast population-averaged brain MRI atlases of a Parkinson’s disease cohort,” accepted in Data in Brief, 2017.
- Y. Xiao, V. Fonov, S. Beriault, F.A. Subaie, M.M. Chakravarty, A.F. Sadikot, G. Bruce Pike, and D. Louis Collins, “Multi-contrast unbiased MRI atlas of a Parkinson’s disease population,” International Journal of Computer-Assisted Radiology and Surgery, vol. 10(3), pp. 329-341, 2015.
- Y. Xiao, S. Beriault, G. Bruce Pike, and D. Louis Collins, “Multicontrast multiecho FLASH MRI for targeting the subthalamic nucleus,” Magnetic Resonance Imaging, vol. 30, pp. 627-640, 2012.
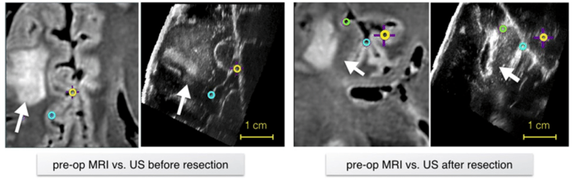
REtroSpective Evaluation of Cerebral Tumors (RESECT) dataset
The repository includes pre-operative MR and intra-operative US images from 23 patients who have received low- grade gliomas resection surgeries (no follow-up re-operations) at St. Olav’s University Hospital. Homologous landmarks are provided between MRI and US, as well as between US volumes during different surgical stages. The database provides a common ground to validate different image registration techniques for improving the surgical treatment of brain cancer. The data are made available with Creative Commons Attribution 4.0 International Public License.
For the methods used, and to use the atlas for research purposes, please cite the following article:
- Y. Xiao, M. Fortin, G. Unsgård , H. Rivaz and I. Reinertsen, “REtroSpective Evaluation of Cerebral Tumors (RESECT): a clinical database of pre-operative MRI and intra-operative ultrasound in low-grade glioma surgeries,” accepted in Medical Physics, 2017.
Instruction for RESECT dataset
- The landmarks of the data are in MINC tag file format with the extension .tag.
- The easiest way to visualize the MINC image pair with the homologous landmarks is to use the software 'register' that comes with MINC Toolkit (https://bic-mni.github.io). To view the images, please use the following command in your terminal:
- Each row of the landmarks with six data entries represent a landmark pair. In each row, the first 3 columns are the x-, y-, and z-coordinate of landmarks belonging to image1 in world coordinates (unit = mm), and the latter 3 columns are the x-, y-, and z-coordinate of landmarks belonging to image2 in world coordinates (unit = mm).
- The reference image is always put as image1. In the RESECT database, this is the MRI or ultrasound before resection.
- The world coordinates of the corresponding MINC and NIFTI files are the same.
EASY-RESECT
EASY RESECT is a simplified sub-set of the original RESECT database. The database contains 22 subjects and intend to help develop MRI vs. US registration algorithms to correct tissue shift in brain tumor resection. All images have been pre-processed and resample to image dimensions that are easy to use for deep learning purposes.
- To facilitate users that are not familiar with medical image data formats, header transformations that links image space and real-world coordinates have also been provided for both NIFTI and MINC formats.
- Anatomical landmarks have been prepared in both formats of coordinates and voxelized images (courtesy of Dr. Mattias Heinrich at University of Luebeck)
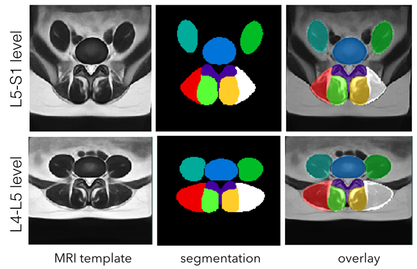
Population-averaged paraspinal muscles atlas
This dataset contains population-averaged T2w MRI atlases of paraspinal muscles at theL4-L5 and L5-S1 levels. Two releases are included in the repository. The 2018 release contains atlases made with left-and-right symmetry while the 2019 release includes asymmetric atlases with anatomical labels.
The atlases were made with 117 patients aged between 30 and 59 years old with commonly diagnosed lumbar pathologies. For atlas construction, the population was divided into three groups: 30-39, 40-49, and 50-59 years old, and for each group,19-21 subjects were included for each of male and female cohorts.
All images are provided in NIfTI-1 format. This dataset is released with the CC BY-NC-SA 3.0 license.
To use the dataset, please cite the following article:
The atlases were made with 117 patients aged between 30 and 59 years old with commonly diagnosed lumbar pathologies. For atlas construction, the population was divided into three groups: 30-39, 40-49, and 50-59 years old, and for each group,19-21 subjects were included for each of male and female cohorts.
All images are provided in NIfTI-1 format. This dataset is released with the CC BY-NC-SA 3.0 license.
To use the dataset, please cite the following article:
- Y. Xiao, M. Fortin, M.C. Battie, H. Rivaz, "Population-averaged MRI atlases for automated image processing and assessments of lumbar paraspinal muscles," European Spine Journal, 2018.