CT brain ventricle segmentation via diffusion Schrödinger Bridge
With the challenges in poor soft tissue contrast and a scarcity of well-annotated databases for clinical brain CTs, we introduce a novel uncertainty-aware ventricle segmentation technique without the need of CT segmentation ground truths by leveraging diffusion-model-based domain adaptation.
For further information, please refer to the following article:
- R Teimouri, M. Kersten-Oertel, Y. Xiao, "CT-based brain ventricle segmentation via diffusion Schrödinger Bridge without target domain ground truths," accepted at the 27th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2024.
MedCLIP-SAM: text-based interactive universal medical image segmentation
We propose a novel framework, called MedCLIP-SAM that combines CLIP and SAM models to generate segmentation of clinical scans using text prompts in both zero-shot and weakly supervised settings. To achieve this, we employed a new Decoupled Hard Negative Noise Contrastive Estimation (DHN-NCE) loss to fine-tune the BiomedCLIP model and the recent gScoreCAM to generate prompts to obtain segmentation masks from SAM in a zero-shot setting.
For further information, please refer to the following article:
For further information, please refer to the following article:
- T Koleilat, H. Rivaz, Y. Xiao, "MedCLIP-SAM: Bridging text and image towards universal medical image segmentation," accepted at the 27th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2024.
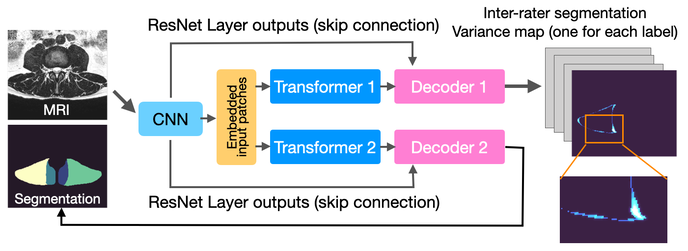
Paraspinal Muscle Segmentation
This repository contains the code for Joint Paraspinal Muscle Segmentation and Inter-rater Labeling Variability Prediction with Multi-task TransUNet. This model is a multi-task TransUNet that provides segmentation masks for paraspinal muscles (left and right erector spinae and multifidus) at the L3-L4,L4-L5, L5-S1, and S1 spinal levels and predicts the pixel-wise variance map of the rater annotations.
For further information, please refer to the following article:
For further information, please refer to the following article:
- P. Roshanzamir, H. Rivaz, J. Ahn, H. Mirza, N. Naghdi, M. Battie, M. Fortin, Y. Xiao, "Joint paraspinal muscle segmentation and inter-rater labeling variability prediction with multi-task TransUNet," the MICCAI workshop on Uncertainty for Safe Utilization of Machine Learning in Medical Imaging (UNSURE), LNCS 13563, pp 125-134, 2022.
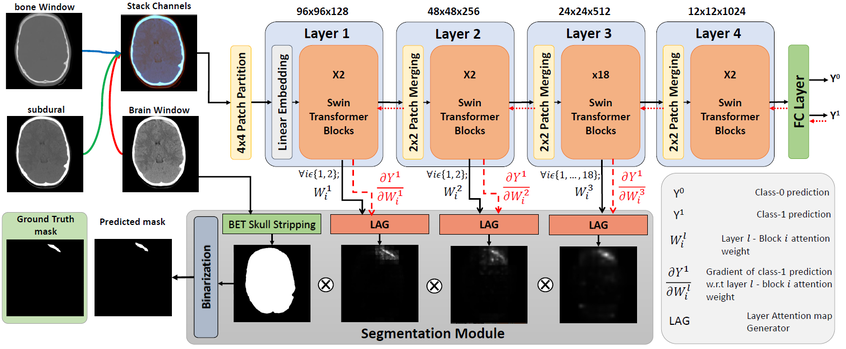
Head-wise Gradient-Infused Self-Attention Map for weakly supervised brain lesion segmentation
This repository contains the code used for the paper: Weakly Supervised Intracranial Hemorrhage Segmentation using Head-Wise Gradient-Infused Self-Attention Maps from a Swin Transformer in Categorical Learning.
For further information, please refer to the following article:
For further information, please refer to the following article:
- A. Rasoulian, S. Salari, Y. Xiao, "Weakly Supervised Intracranial Hemorrhage Segmentation using Head-Wise Gradient-Infused Self-Attention Maps from a Swin Transformer in Categorical Learning," the Machine Learning and for Biomedical Imaging (MELBA) Journal, vol 2, 338-360, 2023.
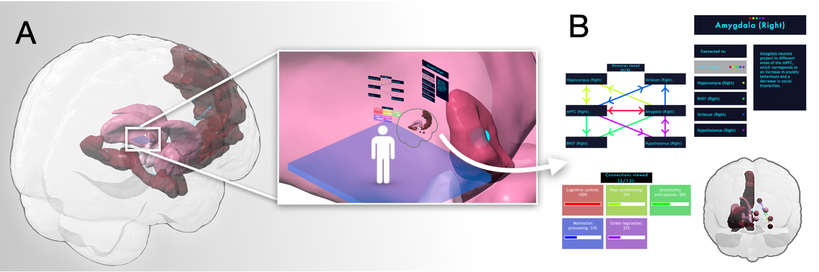
SONIA: immerSive custOmizable Neuro learnIng plAtform
The repository contains the code and assets for the immerSive custOmizable Neuro learnIng plAform (SONIA), a novel user-friendly VR software system with a multi-scale interaction paradigm that allows flexible customization of learning materials. The systems was designed for Virtual Reality (VR) in Unity with SteamVR and the HTC VIVE Pro head-mounted display.
For further information, please refer to the following article:
For further information, please refer to the following article:
- O. Hellum, C. Steele, Y. Xiao, “SONIA: an immersive customizable virtual reality system for the education and exploration of brain networks,” Frontiers in Virtual Reality, 2023.
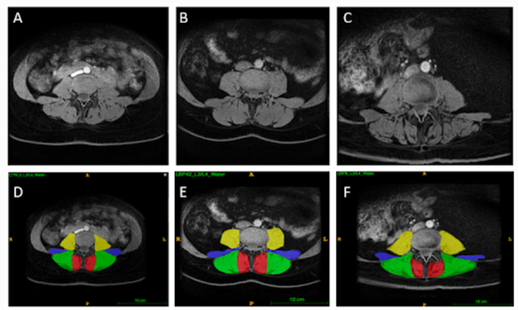
Project PILLAR: a comprehensive online guide for paraspinal muscle segmentation
The ParaspInaL muscLe segmentAtion pRoject (PILLAR) is a comprehensive on-line resource of protocols and tutorials for manual segmentation of paraspinal muscles. The literature on paraspinal musculature is expanding. While this is fantastic news for investigating the morphology and function of these muscles and their relationships with pathologies and biomechanics, there is currently no standardized segmentation protocol. It is imperative to have consistent guidelines in order to properly compare results and further our understanding of the role paraspinal musculature plays in the body and in varying pathologies. The PILLAR project aims to provide new users with easy to follow protocols, video tutorials, instructions, and introduce an open source software for segmentations that is user-friendly.
For further information, please refer to the following article:
For further information, please refer to the following article:
- M. Anstruther, B. Rossini, T. Zhang, T. Liang, Y. Xiao, M. Fortin, "PILLAR: ParaspInaL muscLe segmentAtion pRoject - a comprehensive online resource to guide manual segmentation of paraspinal muscles from Magnetic Resonance Imaging," BMC Musculoskelet Disord 24, 909, 2023.